

To stay up-to-date with the lab, don't forget to follow us on Twitter (@BodenmillerLab)!
News
-
Bernd Bodenmiller discusses cutting-edge cancer microscopy in a recent articleJun 24
For roughly 400 years, microscopes have allowed us to observe increasingly smaller details. Today’s most advanced instruments can peer deep into living cells, helping researchers study diseases such as cancer and improve therapies. Several research groups at UZH are working toward this goal.
Read more here -
Bernd was invited to the Strategy & Insider podcast to talk precision oncology, AI and the future of diagnostics.May 30
Alongside Prof. Dr. Andreas Wicki, Bernd shared his vision to transform cancer diagnosis, treatment, and management. Click the link below to listen to his take on the future of diagnostics.
-
We're pleased to announce two new Ph.D. students, Quentin Hellier and Simone Häfliger. Welcome on board!Jan 8
-
Huge congratulations to Tsuyoshi for successfully defending his PhD!Oct 26
The subject of his Thesis is: "Highly-Multiplexed and Sensitive Antibody-Based Imaging with DNA Barcodes and Metal Isotope-Labeling".
-
A big welcome to our newest post-doctoral fellow, Genki Usui! We're delighted to have you join our team.Oct 2
-
Today, the Bodenmiller lab celebrated its 10 year anniversary. Thanks to all our alumni who travelled far and wide to join us!Sep 8
-
Welcome to our newest team members, Lydia Schulla and Egle Ervin. We're thrilled to have them on board!Sep 1
-
Congratulations to Lena for successfully defending her PhD!Jul 13
The title of her Thesis is: Single-Cell Analysis of Cancer-Associated Fibroblast Heterogeneity with a Focus on
Spatial Distribution and Clinical Implications. Lena's PhD was awarded with a Distinction. -
We're excited to announce the newest member of the team! A big welcome to our new postdoctoral fellow, Maria Ramos Zapatero!Jul 3
-
A warm welcome to our new PhD student, Nathan Steenbuck!Apr 3
-
A big welcome to our new senior computational scientist, Milad Adibi and our new research associate, Sophie Déglise!Feb 1
-
We're excited to open our multiplexed tissue imaging workshop to a wider audience! For more details, click here.Dec 8
Given high demand, the multiplexed imaging symposium and parts of the computational workshop will now also be offered online. Register with the QR code here and check out the attached details.
- For a more detailed overview of the symposium, see our programme here.
- For details of the computational workshop, see our overview here.
-
We are pleased to announce our multixed tissue imaging workshop! Get hands-on experience with multiplexed imaging and hear from experts in the field. Click here for more details.Oct 25
Our workshop takes place in Zurich from the 09 - 12 January, 2023, offering a comprehensive dive into the field of multiplexed imaging. In-person tickets are currently sold out.
-
Jonas successfully defended his PhD!Jun 22
The subject of his Thesis is: “ Multiplexed Tissue Image Processing and Spatial Single-Cell Data Integration”. Congratulations to Jonas!
-
Laura successfully defended her PhD!Mar 29
The subject of her Thesis is: “Studying Breast Cancer Invasion and Metastasis with 2D and 3D Imaging Mass Cytometry ”. Congratulations to Laura!
-
Bernd was a guest on the Strategy & Insider podcast with Dr. Thomas SolbachDec 2
Algorithms could dramatically revolutionize healthcare and modern medicine by making predictions about cancer treatments and providing patients with personalized and individual care. Together with Dr. Thomas Solbach, Bernd discussed how digitization could spur healthcare research to the next level in the latest Strategy&Insider podcast episode.
-
Jana awarded for outstanding doctoral thesisDec 1
Jana R Fischer has received a distinction for outstanding dissertation by the Faculty of Science on the 15th of November 2021.
The subject of her Thesis is: “High-Dimensional Profiling of the Breast Cancer Microarchitecture”. Congratulations to Jana!
-
Sunny successfully defended her PhD!Nov 1
The subject of her Thesis is: “Single-Cell Profiling of the Tumor Immune Microenvironment of Primary and Metastatic Human Breast Cancer”. Congratulations to Sunny!
-
Jana successfully defended her PhD!Oct 2
The subject of her Thesis is: “High-Dimensional Profiling of the Breast Cancer Microarchitecture”. Congratulations to Jana!
-
New members in the Bodenmiller lab.Oct 1
Stefanos Voglis has joined the lab as a visiting scientist and Alina Bollhagen has started as a Ph.D. student.
-
New members in the Bodenmiller lab.Jan 4
Pierre Bost has joined the lab as a postdoctoral fellow, Tatjana Schmitz and Mengze Zhang have started as Ph.D. students, and Eduard Petrosyan has joined us as a Master's student.
-
Bernd Bodenmiller has been appointed dual professor.Sep 20
Bernd Bodenmiller is now dual professor at the Department of Quantitative Biomedicine, UZH and at the Institute for Molecular Health Sciences, ETH Zurich.
-
New students in the Bodenmiller lab.Sep 1
Daria Lazic is a visiting Ph.D. student, learning to apply imaging mass cytometry in the Bodenmiller lab. Luca Räss and Hangjia Zhao have joined us as Master's students.
-
Johanna received the “Annual Award 2020” of the Faculty of Sciences for her PhD ThesisApr 28
Auf Antrag der Mathematisch-naturwissenschaftlichen Fakultät verleiht die Universität Zürich einen Jahrespreis an
Dr. Johanna Wagner-Albrecht für ihre Dissertation «Single-Cell Proteomic Characterization of the Tumor and Immune Ecosystem of Human Breast Cancer with Focus on Metastatic Potential».
Johanna erforscht die Diversität der Zellen in humanen Brusttumoren. Sie entdeckte neue Patientinnen Untergruppen die von Immuntherapien profitieren könnten und dass aggressivere Tumore überraschenderweise von wenigen Krebszellarten dominiert werden. Ihre Arbeit ist richtungsweisend für individuell auf Patientinnen zugeschnittene Therapien.
Congratulations Johanna! -
Marco successfully defended his PhD!Jan 21
The subject of his Thesis is: “Deciphering the Signaling Network Landscape of Breast Cancer Supports Precision Medicine”. Congratulations to Marco!
-
Vito successfully defended his PhD!Jan 7
The subject of his Thesis is: “Investigation of Intra- and Intercellular Signaling through Mass Cytometry Based Single Cell Methods”. Congratulations to Vito!
-
Johanna awarded for outstanding doctoral thesesJan 7
Johanna Wagner has received a distinction for outstanding dissertation by the Faculty of Science on the 13th of December 2019. That means that Johannas’ thesis was among the top 5% PhD theses awarded by the University of Zurich, based on a jury of international reviewers.
The subject of her Thesis is: “Single-Cell Proteomic Characterization of the Tumor and Immune Ecosystem of Breast Cancer”. Congratulations to Johanna!
-
Bernd and his team have been awarded with the ERC Consolidator GrantDec 16
Tumors are highly complex entities that consist of many different cells communicating with each other. The project aims to develop new technologies and computer-aided methods that rationalize this complexity and describe tissues akin to (a)social networks. Such representations could help scientists understand the mechanistic underpinnings of cancer in the context of metastatic breast cancer and determine the most suitable therapies for breast cancer patients.
Congratulations!
To stay up-to-date with the lab, don't forget to follow us on Twitter (@BodenmillerLab)!
Research
Methods development
We develop experimental and computational methods to study tumor ecosystems on the single-cell level. In such an ecosystem, many cell types, including tumor, stromal, immune and endothelial cells, interact and communicate in multicellular assemblies. We seek to understand how tumor ecosystems function and ultimately how their properties affect disease. We generate comprehensive single-cell datasets of tumors from many patients, which requires detection of dozens of markers simultaneously. We have pioneered mass cytometry-based methods for simultaneous multiplexed marker detection and analysis in both dissociated tissues and on tissue sections, and continue to develop these methods further (see technology section).
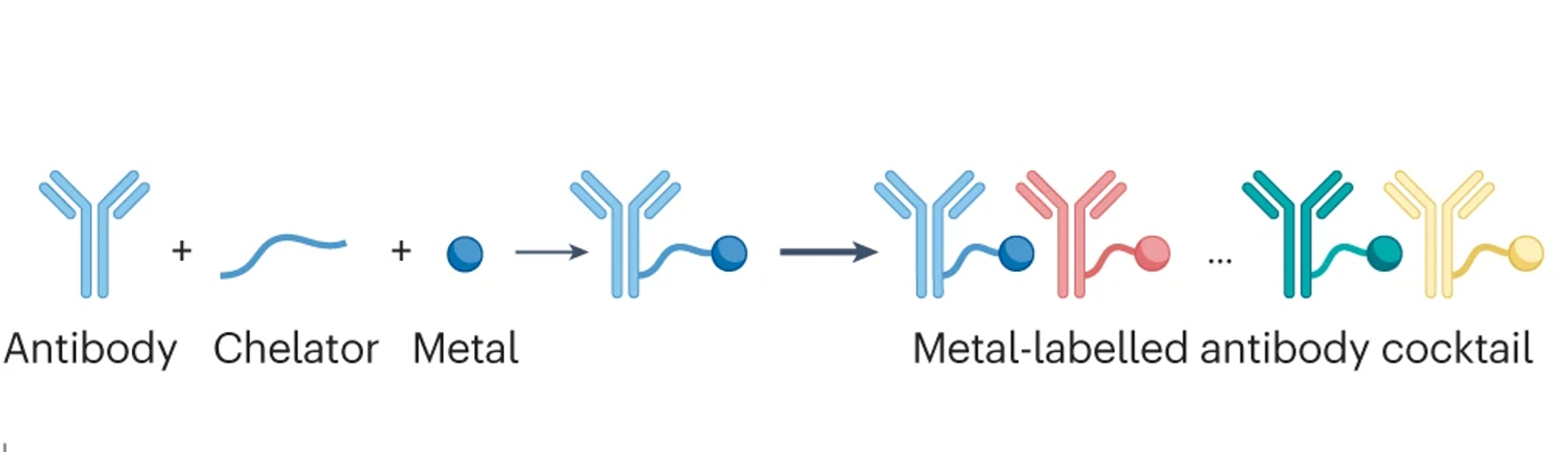
de Souza et al., 2024 Nat. Reviews Cancer
Translational research and precision medicine
Solid tumors are multicellular ecosystems of diverse cell types that interact to manifest emergent phenotypes which ultimately determine clinical outcome. We combine suspension or imaging mass cytometry with computational techniques to systematically describe cell phenotypes in tumor ecosystems and to examine their distribution across patients and tumor types. We generate atlases of human tumors from large patient cohorts with known clinical outcome, and identify cellular and spatial phenotypes associated with disease progression. These atlases lay the foundation for improved patient stratification and provide potential drug targets for ongoing study. These data are also the foundation for follow-up studies to understand mechanisms of the disease. We are part of several large-scale, multi-center projects bringing together clinicians, research labs and pharmaceutical companies, in which we develop mass cytometry and imaging mass cytometry for precision medicince applications.

Artistic representation of an invasive breast tumor ecosystem.

Image by Johanna Wagner.
Mechanisms of cancer
Single-cell systems biology analyses of tumor samples yield a wealth of data about cancer biology. To understand the regulatory networks underlying the disease, we use algorithmic and data-driven approaches to model subpopulations of cells and their signaling network structures. Further, using data from imaging mass cytometry, we model how signaling network states spatially couple with those of other cells. As a complement to modeling and associative studies, we use in vitro patient-derived organoid and cell co-culture models to conduct small molecule screens and carry out perturbation studies aimed at understanding mechanistic aspects of tumor biology.
Breast cancer organoid cultures
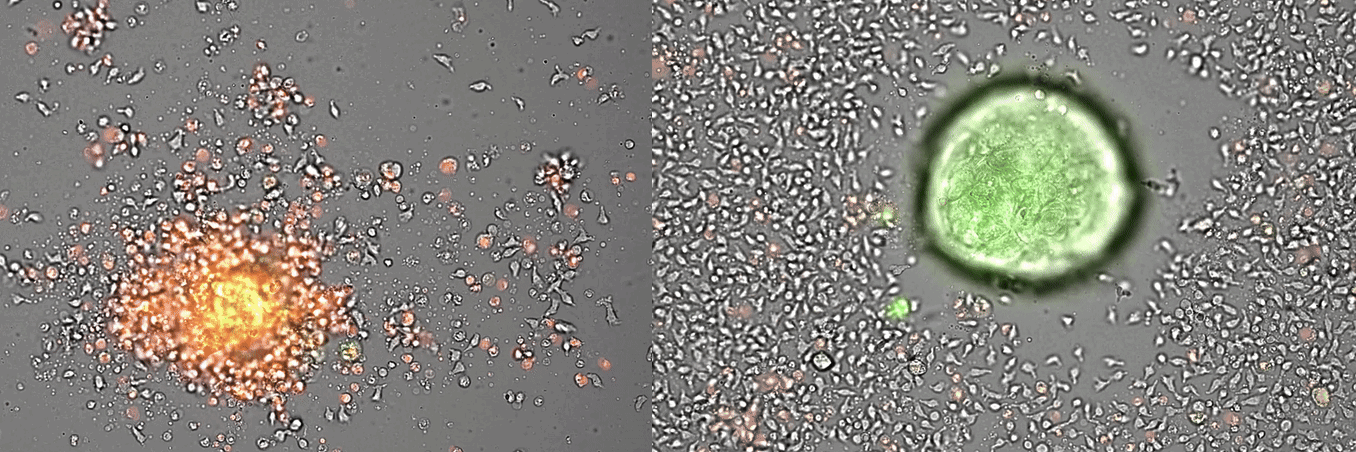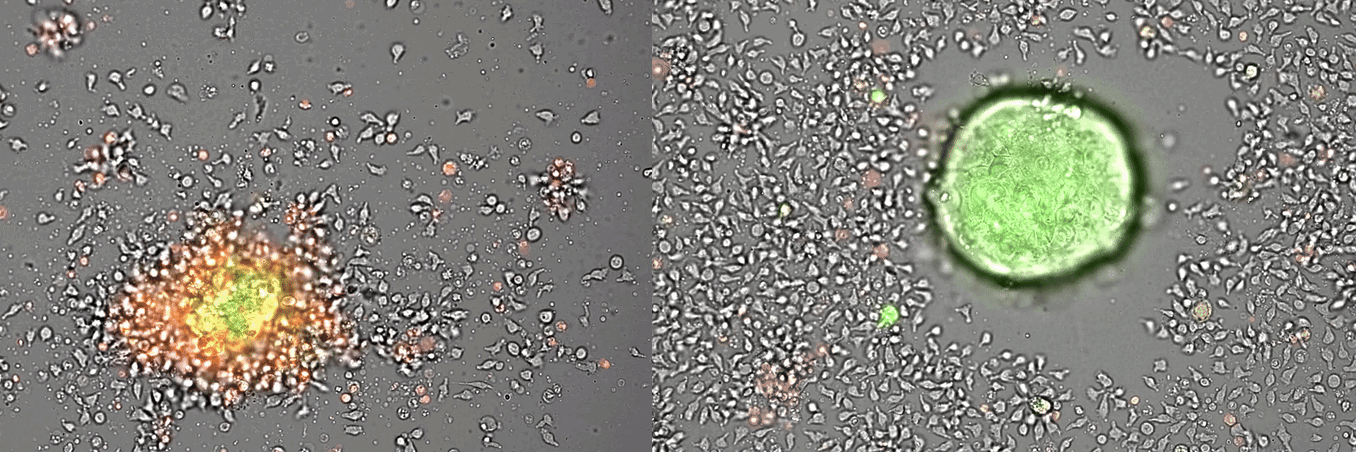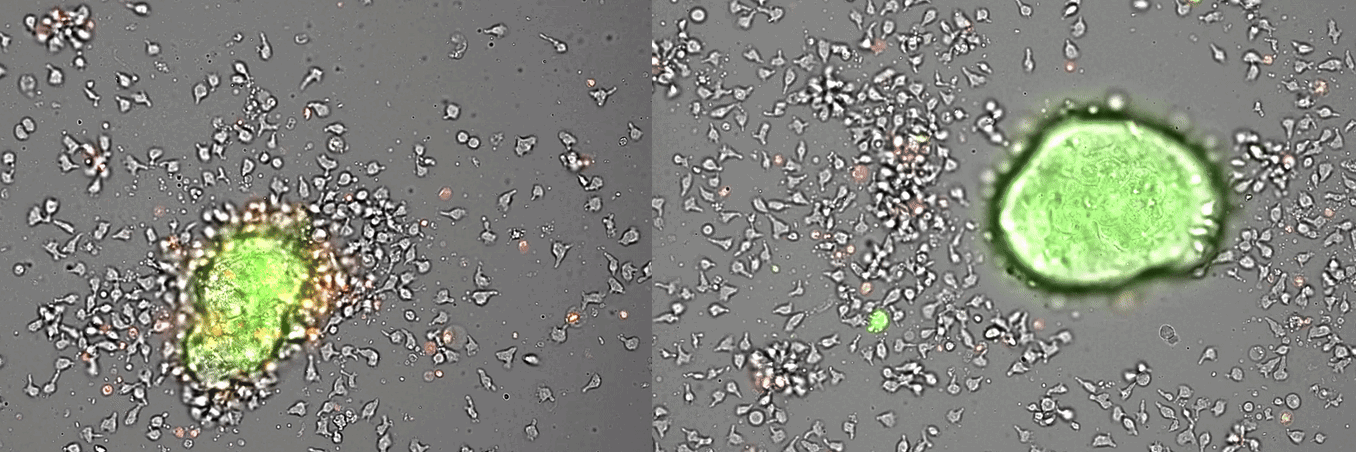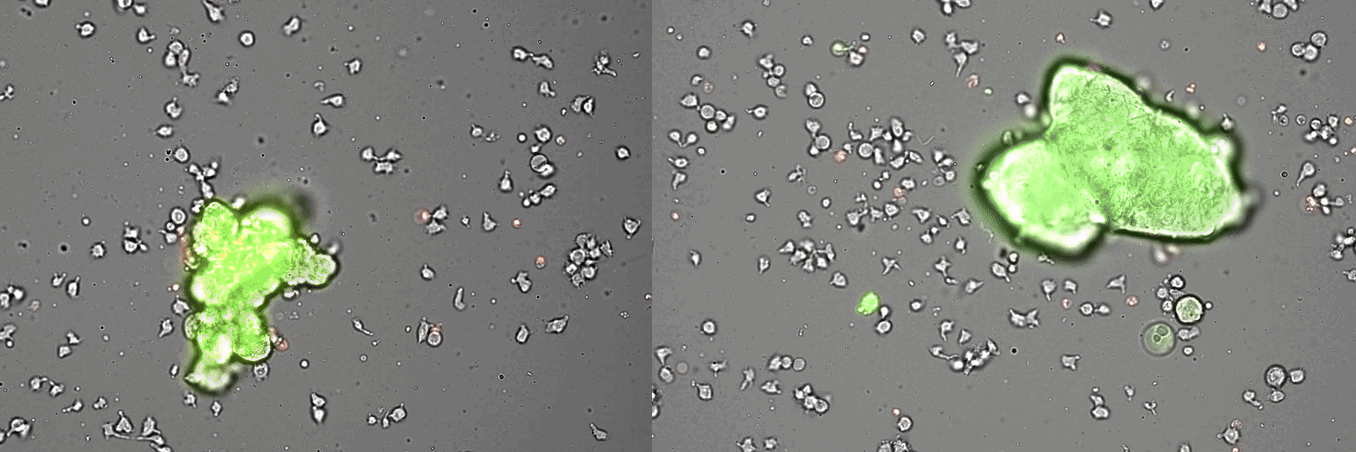
(courtesy of Tatjana Schmitz)
Selected Publications
Technology
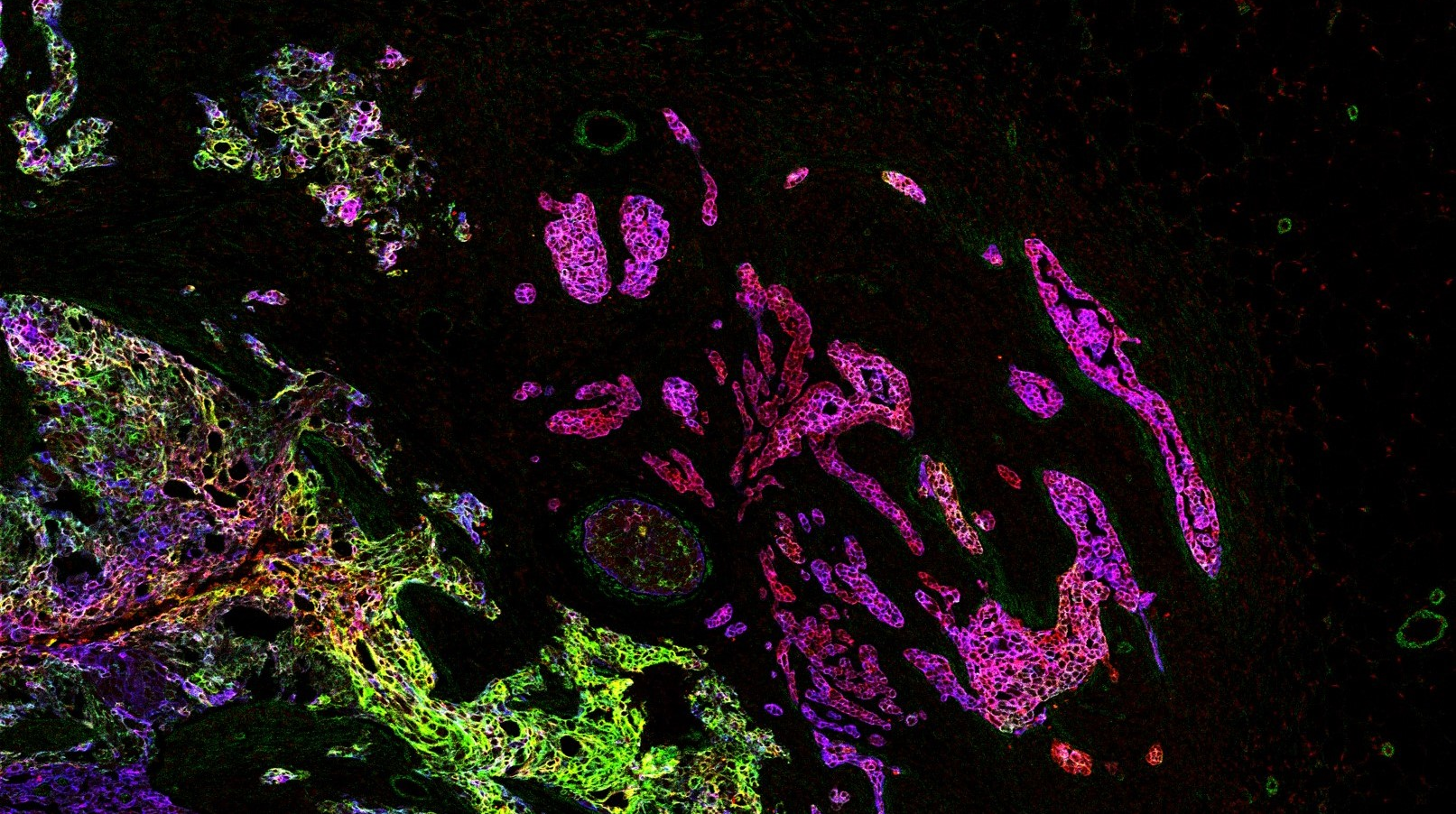
Imaging Mass Cytometry
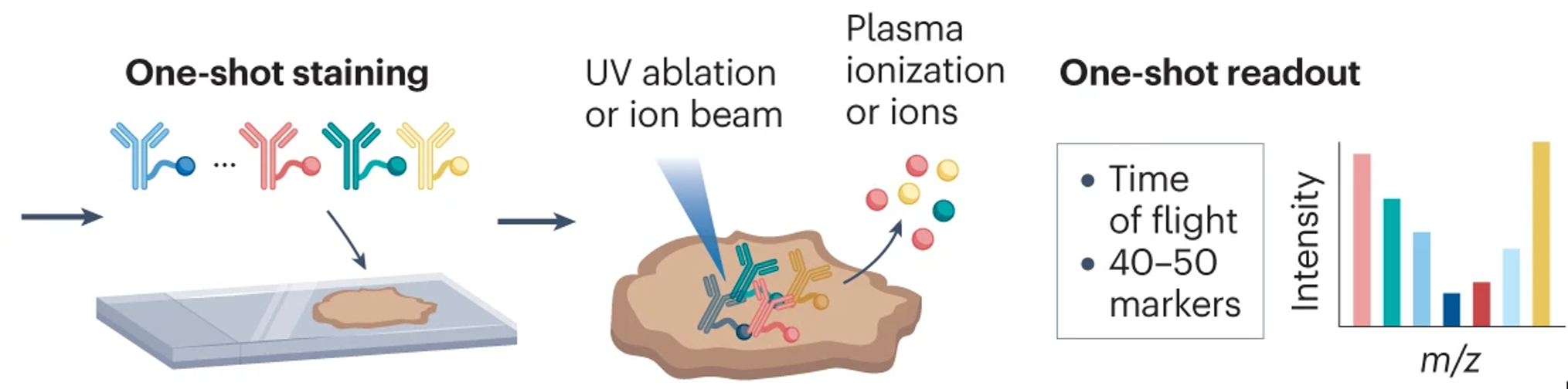
Single-cell systems biology of cancer requires methods to measure multiple markers within tumors, quantitatively, and with spatial and single-cell resolution. Based on our earlier work on suspension mass cytometry, the Bodenmiller group has pioneered a spatial mass spectrometric approach called imaging mass cytometry (IMC) for the simultaneous and spatially-resolved quantification of approximately 50 markers on single cells. We employ IMC-based methods to study the cellular composition, spatial organization and regulation of tissue ecosystems, for insights into health and disease.
Imaging mass cytometry: measurement
In mass cytometry, we use a mass spectrometer to measure protein and/or transcript levels within cells, using antibodies or RNA probes linked to different metal isotopes. In imaging mass cytometry, we have extended this technology to solid tissue samples such as tumor biopsies, analyzing them spatially and capturing effects of the local microenvironment on tumor cells. We apply IMC in 2D and are also developing it in 3D. Mass cytometry can in principle reliably differentiate over a 100 probes. We are continuously improving the speed, number of markers, resolution, reliability, sensitivity, biological interpretability and overall quality of high-dimensional single cell analysis.
Imaging mass cytometry: analysis
Deriving relevant biological information from high-dimensional datasets is an ongoing challenge in systems biology. We explore the capabilities of existing statistical and image analysis tools to analyse imaging mass cytometry data in a meaningful way. We also develop software to process, visualize and analyze high dimensional IMC datasets.


Software
The Bodenmiller GitHub page has code and scripts for many projects. The IMC workflow provides an overview on IMC data analysis approaches.
histoCAT


histoCAT is an open-source visualization and analysis toolbox for exploration of rich multidimensional IMC datasets. It enables parallel visualization of images and single cell phenotypic distributions and includes methods to identify and quantify cell-cell interactions within tissue. Read the paper.
steinbock


steinbock is a collection of tools for multi-channel image processing using the command-line or Python code. Supported tasks include IMC data preprocessing, supervised multi-channel image segmentation, object quantification and data export to a variety of file formats. The steinbock framework is fully documented, integrates well with downstream analysis packages and is available as platform-independent Docker container, ensuring reproducibility.
cytomapper


Multiplex imaging cytometry acquires spatially-resolved single-cell expression values of selected proteins in a sample. Cytomapper can be used to visualize the multiplexed read-outs obtained with this technique. The main functions of this package allow (i) the visualization of pixel-level information across multiple channels and (ii) the display of cell-level information (expression and/or metadata) on segmentation masks.
imcRtools


The imcRtools R/Bioconductor package supports the handling and analysis of imaging mass cytometry and other highly multiplexed imaging data. The main functionality includes reading in single-cell data after image segmentation and measurement, data formatting to perform channel spillover correction and a number of spatial analysis approaches.
Protocols
The Bodenmiller lab protocols page is in progress.
People
Current Members
Prof. Dr.
Single-cell systems biology of cancer requires methods to measure multiple markers within tumors, quantitatively and at single-cell resolution. To this end, the Bodenmiller group has pioneered a single-cell mass spectrometric approach called mass cytometry (CyTOFTM). This technology allows the simultaneous and high-throughput quantification of approximately 50 markers, including proteins and their modifications, on single cells. We employ mass cytometry-based methods to study the cellular composition and regulation of tissue ecosystems, for insights into health and disease.
Laboratory Manager
Senior Research Associate
Alumni
-
Miriam Rüfenacht , B.Sc.
Master Student -
Milad Adibi, Ph.D.
Roche AG -
Raza Ali, M.D./Ph.D.
Group Leader, CRUK Cambridge Institute -
Maya Barben, Ph.D.
ETH Zürich -
Pierre Bost, Ph.D.
Marie Curie Institute -
Marcel Burger, Ph.D.
Wissenschaftlicher Mitarbeiter Elemental Analysis, Solvias -
Ruben Casanova, Ph.D.
Senior Data Scientist -
Raúl Catena, Ph.D.
Senior Software Engineer, Leica Geosystems -
Stephane Chevrier, Ph.D.
Co-founder, Navignostics AG -
Lena Cords, Ph.D.
Postdoctoral Fellow - TU München -
Haithem Dakhli, Ph.D.
-
Nicolas Damond, Ph.D.
Postdoctoral Fellow -
Esther Danenberg, M.Sc.
Research Administrator, CRUK Cambridge Institute -
Michelle Daniel, M.Sc.
Navignostics AG -
Nadine Dobberstein, M.Sc.
Technician, InterAx Biotech -
Nils Eling, Ph.D.
Senior Data Scientist, Novartis -
Bruno Palau Fernandez, B.Sc.
Research Assistant -
Jana Fischer, Ph.D.
Co-founder, Navignostics AG -
Catrina Friedrich, M.Sc.
Master's Student -
Charlotte Giesen, Ph.D.
Head, Quality Assurance, Roche -
Tobias Hoch, M.Sc.
Research Associate, Empa -
Tsuyoshi Hosogane, Ph.D.
Postdoctoral Fellow -
Alexandra Huber, B.Sc.
Master's student, ETH Zurich -
Hartland Jackson, Ph.D.
Group Leader, Lunenfeld-Tanenbaum Institute -
Andrea Jacobs
Co-founder, Navignostics AG -
Merel Kuijs, M.Sc.
Genbio AI -
Laura Kütt, Ph.D.
Ph.D. -
Pieter Langerhorst, M.Sc.
Ph.D. Student, Radboud Institute -
Daria Lazic, M.Sc
Post-doctoral fellow -
Xiaokang Lun, Ph.D.
Postdoctoral Fellow, Wyss Institute -
Constance Lyon, M.Sc.
Current affiliation unknown -
Markus Masek, M.Sc.
Ph.D. Student, University of Zurich -
Alaz Özcan, M.Sc.
Ph.D. Student, University of Zurich -
Serena Di Palma, Ph.D.
Assistant Professor, Utrecht University -
Adhvitha Premanand, B.Tech
-
Swetha Raghuraman, M.Sc.
Ph.D. Student, University of Muenster -
Luca Räss, M.Sc.
Scientist R&D Automatization, Biognosys AG -
Anton Rau, M.Sc.
Software Engineer -
Leonor Schubert Santana, M.Sc.
Master's Student -
Denis Schapiro, Ph.D.
Postdoctoral Fellow, Broad Institute -
Yannik Severin, M.Sc.
Ph.D. Student, ETH Zurich -
Sujana Sivapatham, M.Sc.
Research Associate, Immunos Therapeutics -
Merrick Strotton, Ph.D.
Prinicpal Scientist, high-plex imaging, UCB -
Sandra Tietscher, Ph.D.
Business Development Manager Sensor Innovation at Sensirion -
Marco Tognetti, Ph.D.
Senior Scientist, Biognosys AG -
Sophie Tritschler, M.Sc.
Ph.D. Student, Helmholtz Z., Muenchen -
Eleni Tselempi, M.Sc.
Senior Research Associate, Roche Innovation Centre -
James Wade, Ph.D.
QSP Expert, LYO-X GmbH -
Johanna Wagner, Ph.D.
Postdoctoral Fellow, NCT Heidelberg -
Jonas Windhager, Ph.D.
Bioimage analyst, SciLife Lab -
Shuhan Xu, M.Sc.
Ph.D. Student, MPI -
Vito Zanotelli, Ph.D.
Data Analytics Consultant, D ONE -
Mengze Zhang, Ph.D.
Graduate Rotation Trainee, Astra Zeneca -
Shan Zhao, Ph.D.
Group Leader -
Nevena Zivanovic, Ph.D.
Research Scientist, Janssen Pharmaceuticals
Co-founder, Navignostics AG
Drug development in breast cancer suffers from a lack of faithful models able to recapitulate breast cancer heterogeneity at the single-cell level. To overcome this limitation, we have generated a patient-derived breast cancer organoid biobank from fresh and frozen breast cancer samples. We are using this model to study the different populations composing the tumor and their response to drug treatments. It is our conviction that the study of the different cellular populations composing breast tumors, and their interactions, will lead to a better understanding of the tumor ecosystem and to the identification of therapies targeting specific cellular populations that could trigger tumor demise.
Postdoctoral Fellow
I am combining highly multiplexed imaging, data analysis and experimental biology to characterize type 1 diabetes (T1D) progression and beta cell heterogeneity.
I did my PhD with Prof. Pedro Herrera at the University of Geneva, studying regeneration of insulin-producing beta cells by transdifferentiation of the closely related alpha cells. During my thesis, I used confocal microscopy and lineage tracing in complex transgenic mouse models and primary human pancreatic islets. This work sparked my interest into both T1D research and image analysis of single cells, two elements that are still at the center of my current projects.
In the Bodenmiller lab, I use imaging mass cytometry to analyze pancreas sections of individuals with or at risk for type 1 diabetes. The multiplexing capacity of this technology enables deep phenotyping of beta cells and of infiltrating immune cells, and mapping of their interactions. The goal of this project is to gain a better understanding of T1D development in the pancreas.
In parallel, I'm combining primary human islet culture with imaging mass cytometry to study how beta cell subpopulations respond to external stimuli relevant to T1D. The aim is to identify beta cell subsets that are more sensitive to destructive signals or, conversely, more responsive to regenerative cues. This work is supported by a JDRF postdoctoral fellowship.
Selected Publications
Research Administrator, CRUK Cambridge Institute
Senior Data Scientist, Novartis
My current research focuses on understanding the molecular changes over breast cancer organoid growth. I'm integrating imaging mass cytometry (IMC) with single-cell based statistical approaches to model spatial heterogeneity in organoids. As part of handling IMC data, I develop software for image and single-cell analysis.
In the past, I have completed my PhD in the Marioni group at the European Bioinformatics Institute and the CRUK Cambridge Institute at the University of Cambridge.
Link to website: nilseling.github.io
Link to Google Scholar: https://scholar.google.com/citations?user=kBIvrFoAAAAJ&hl=de
Link to Github: https://github.com/nilseling
Co-founder, Navignostics AG
Research Associate, Empa
As a research assistant and former Master's student in the lab, I am currently analyzing IMC data with a focus on the immune system and its relation to the protein family of chemokines. I am investigating whether and to what extent the spatial context of chemokine expression can contribute to understanding differences in the immune landscape of patients.
Group Leader, Lunenfeld-Tanenbaum Institute
Co-founder, Navignostics AG
Business Development Manager Sensor Innovation at Sensirion
Postdoctoral Fellow, NCT Heidelberg
Selected Publications
Data Analytics Consultant, D ONE
I like to develop high throughput methods together with tailored data analysis approaches to better understand how cells perceive their environment and interact.
Lab Life
Open Positions
We are always looking for excellent and motivated students/postdocs with a strong background in biological/biochemical/biomedical research or bioinformatics.
Prospective Ph.D. students can apply to our group via the Life Science Zurich Graduate School. Our group is a member of the Molecular Life Sciences, the Systems Biology and the Cancer Biology programs. For computational students, we offer shared Ph.D. positions with bioinformatics research groups.
Post-doctoral candidates should be highly motivated with a passion for science with great interest in quantitative biology, single-cell analysis and systems biomedicine. Our lab and collaborators provide an excellent and vibrant interdisciplinary environment, including systems (cancer) biology, biochemistry, analytical sciences, computer sciences and biomedical research. Candidates should have a demonstrated record of productivity during their Ph.D. studies, including publications in peer-reviewed journals (see DORA).
Applications should be directly sent to Bernd Bodenmiller, including your CV, Publication List and a short paragraph with your scientific interests and what you hope to achieve during your postdoctoral time.
All Publications
For an up-to-date list of our publications, please search PubMed here.
Teaching
We provide a range of master and semester projects for motivated students with interest in biological/biochemical/biomedical research or bioinformatics. Applications should be sent directly to Bernd Bodenmiller, including your CV and a short paragraph with your scientific interests.
Alternatively, for hands-on experience in the Bodenmiller lab, we offer the block courses BME331 & BME330 in the Spring and Autumn semester respectively.
For details of provided lectures, please see course descriptions at UZH and ETHZ.
Computational Biology Master Thesis Project in the Bodenmiller lab
Background
The Bodenmiller lab is part of the “IMMUcan” consortium which set out in 2019 to profile the tumor microenvironment of 2500 cancer patients with multiplexed imaging technics, bulk RNAseq, whole exome sequencing (WES) and hematoxylin & eosin (H&E) staining. As part of this project, we are investigating a patient cohort of advanced-stage non-small cell lung cancer (NSCLC). While these patients typically receive immunotherapy treatment, the response rates are poor and most patient eventually die within 5 years of diagnosis.
Project
For this project we collected multiple data types from up to 210 patients including imaging mass cytometry, whole-slide multiplexed immunofluorescence, H&Es, RNAseq and WES. Using this rich dataset, we want to identify predictors of treatment response for each data type, and using all data types together. Some specific tasks of the project encompass:
- Applying pre-trained deep learning methods (e.g. UNIv2, TITAN) for feature extraction from H&E images (e.g. https://github.com/mahmoodlab/TITAN)
- Train multiple-instance learning model to identify purely H&E based predictive features (e.g. https://github.com/mahmoodlab/CLAM/tree/master)
- Apply Hook-netTLS to identify Tertiary lymphoid structures (TLS) in H&Es (https://github.com/DIAGNijmegen/pathology-hooknet-tls) and calculate summaries and comparison of detected TLS across samples.
- Use block forest or similar to identify most predictive features for treatment response across the whole dataset.
- Depending on time and progress, exploratory data analysis using the complete data set and a validation data set are possible.
What We Offer
- A dynamic, fun and interdisciplinary research environment at the intersection of machine learning and biological sciences
- The chance to work closely with leading researchers in computational biology and biomedical data science
- Hands-on experience applying deep learning to biomedical research
- A unique dataset to investigate biomarkers for the treatment of NSCLC
Who Should Apply?
- Self-driven, motivated students with a background in computational biology or bioinformatics
- Starting date: no later than March 2026
- Proficient in Python, PyTorch and ideally also R
- Previous experience in cancer biology is a plus
How to apply
Interested students should submit the following documents:
- CV (max. 2 pages)
- A short motivation letter (max. 1)
- Transcript of records (we will focus primarily on relevant coursework rather than overall grades)
- Optional: A link to a GitHub repository showcasing a relevant application
- Applications should be sent to daniel.schulz@uzh.ch
Experimental Master’s thesis opportunity: High-throughput drug screening in breast cancer organoids
Breast cancer is the leading cause of cancer-related mortality in women with approximately 670,000 deaths worldwide in 2022. Thus, therapies against breast cancer remain ineffective for many patients, largely due to cancer cell heterogeneity. This project offers a unique opportunity to work with high-throughput drug screening data and to experimentally validate drug response hits. You will gain hands-on experience with cutting-edge 3D culture systems, perform viability assays, and carry out advanced single-cell analyses using mass cytometry (CyTOF). The long-term goal of this work is to develop precision oncology approaches that target cancer cell heterogeneity.
What we are looking for
We are seeking a motivated, organised and reliable Master’s student with a strong interest in cancer biology and experimental lab work. Previous cell culture experience, eagerness to learn new techniques, and the ability to work both independently and collaboratively are essential.
What we offer
We offer training in 3D organoid culture, drug screening and mass cytometry, along with close supervision and a supportive team atmosphere.
Please send your short motivational letter, transcript of records and CV to Egle Helene Ervin (postdoctoral researcher, eervin@ethz.ch)
Master Thesis Project Proposal: Optimization of region selection for spatial proteomics through multimodal analysis of cancer tissue.
Spatial proteomics provides unprecedented insights into the tumor microenvironment and reveals actionable insights enabling personalized cancer treatment. Yet, due to tradeoffs between spatial scale, resolution and multiplexing ability in currently available methods, the results of spatial proteomics analysis depend on manual choices made during the used analysis workflows. Imaging mass cytometry (IMC) provides high-plex single cell information, but on large tissue sections data acquisition is limited to regions of interest (ROIs). Currently, informative ROIs are chosen by a trained pathologist using visual inspection of H&E images (single cell, but low-plex) and low-resolution whole slide IMC images (high-plex but not single cell). We aim to design an algorithm that integrates data from the two complementary modalities and chooses relevant and representative ROIs in a robust and interpretable way. The project will cover the following components:
- Feature extraction from H&E images using deep learning with state-of-the-art digital pathology foundation models
- Feature extraction from low-resolution whole slide IMC images and feature aggregation across modalities
- Development of metrics to measure ROI relevance and representativeness
- Algorithm design for selecting ROIs that optimize the developed metrics
The project will be jointly supervised by Victor Ibañez (Computational Biologist @ Bodenmiller Lab, UZH & ETH) and Benjamin Ehret (Computational Manager @ Navignostics AG). Interested applicants, please send a CV, transcript of records and short motivation letter (max. 400 words) to victor.ibanez@uzh.ch and benjamin.ehret@navignostics.ch. Students with a strong background in computer vision and deep learning are preferred. Minimal project duration is 6 months.
Comptational Master Thesis Project Proposal: Deep Learning for Oncology
Master’s Thesis or Other Credit-Earning Assignment in Deep Learning for Oncology
The Bodenmiller Lab at UZH/ETH is seeking talented and motivated Master’s students with expertise in machine learning to join our team. We offer exciting opportunities to work on cutting-edge deep learning projects focused on tumor-tissue image analysis as part of a Master’s thesis or a credit-earning assignment (e.g. unpaid internship, lab rotation, semester project).
About the Projects
Imaging Mass Cytometry (IMC) is an advanced imaging technology that combines mass spectrometry with microscopy to generate highly multiplexed spatial data at single-cell resolution. Unlike traditional imaging methods that rely on fluorescent markers, IMC uses metal-tagged antibodies to detect dozens of proteins simultaneously within a tissue sample. This enables researchers to study complex cellular environments—such as tumors or immune tissues—with exceptional detail. Currently, the standard approach for analyzing IMC images of tumor tissue involves cell segmentation, where individual cells are identified and treated as the smallest units of analysis. However, this method introduces potential biases and overlooks valuable contextual information in the surrounding tissue. To overcome these limitations, we are developing deep learning approaches that analyze IMC data directly at the pixel level, enabling a more comprehensive and unbiased understanding of tissue architecture and cellular interactions. Potential Projects:
- Optimization of Vision Transformer (ViT) architectures to improve IMC image data analysis
- Enhancing explainability of pixel-based IMC models to better understand deep learning predictions
- Validating pixel-based IMC models by comparing them to single-cell analysis techniques
These projects offer a unique opportunity to apply and develop advanced AI models, work with high-dimensional biological data, and contribute to impactful biomedical research.
What We Offer
- A dynamic and interdisciplinary research environment at the intersection of machine learning and biological sciences
- The chance to work closely with leading researchers in computational biology and biomedical data science
- Hands-on experience applying deep learning to biomedical research
- Flexible start dates
Who Should Apply?
We are looking for students who:
- Are proficient in Python and PyTorch
- Are self-reliant and have strong problem-solving skills
- Have an interest in computer vision and cancer biology
Nice to have:
- A background in computer science, engineering, computational biology or bioinformatics
- Software or machine learning engineering experience
- Experience with high-performance computing environments
- Experience with workflow management systems
How to Apply
Interested students should submit the following documents:
- CV (max. 2 pages)
- A short motivation letter (max. 1)
- Transcript of records (we will focus primarily on relevant coursework rather than overall grades)
- Optional: A link to a GitHub repository showcasing a relevant application
Applications should be sent in ONE email addressed to both
Contact
Department of Quantitative Biomedicine
University of Zurich
Winterthurerstrasse 190
CH-8057 Zurich
Switzerland
We are not easy to find, download our campus map.
Building / Room: Y38-M
phone: +41 44 635 48 25
fax: +41 44 635 68 79
Inst. f. Molecular Health Sciences
ETH Zurich
Otto-Stern-Weg 7
CH-8093 Zürich
Switzerland
Building / Room: HPL H 16.2
phone: +41 44 633 29 20